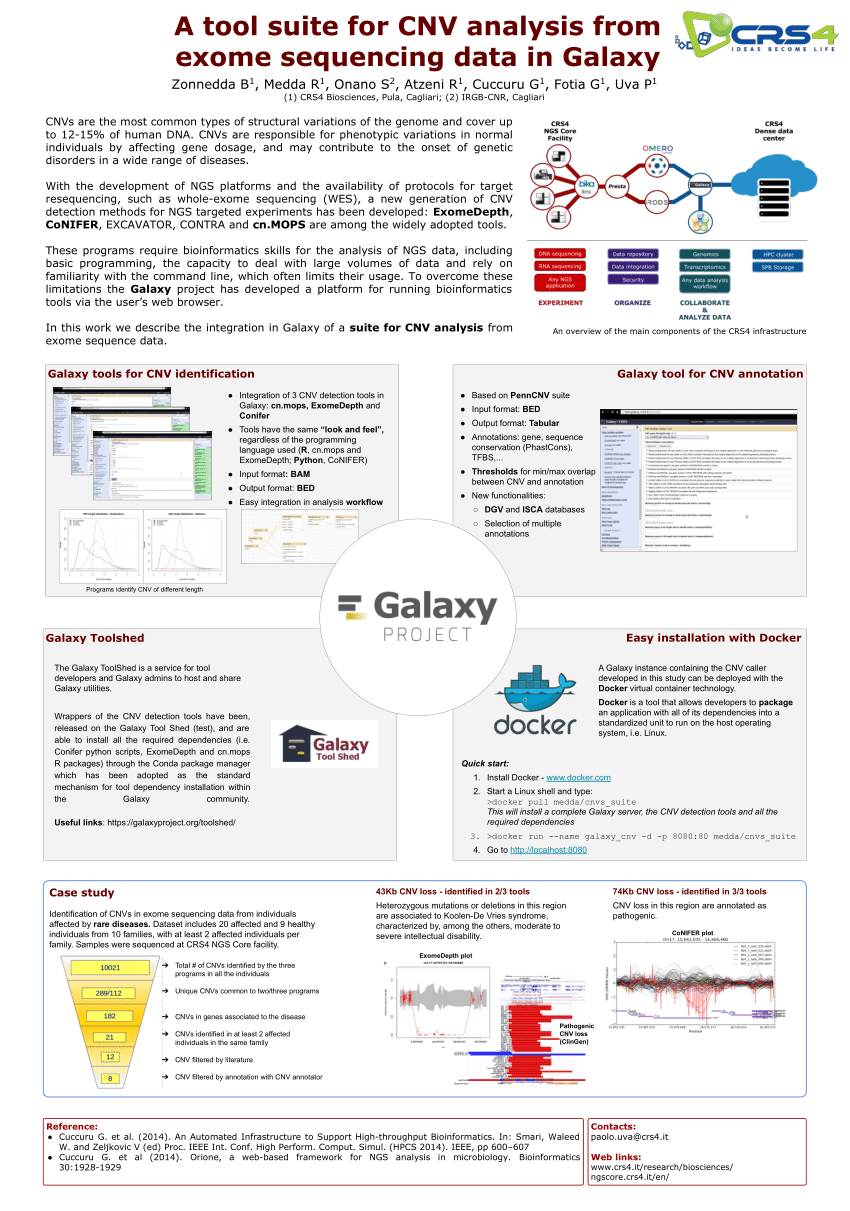
The changes of tools' performances with respect to the CNV size. Fig a... | Download Scientific Diagram
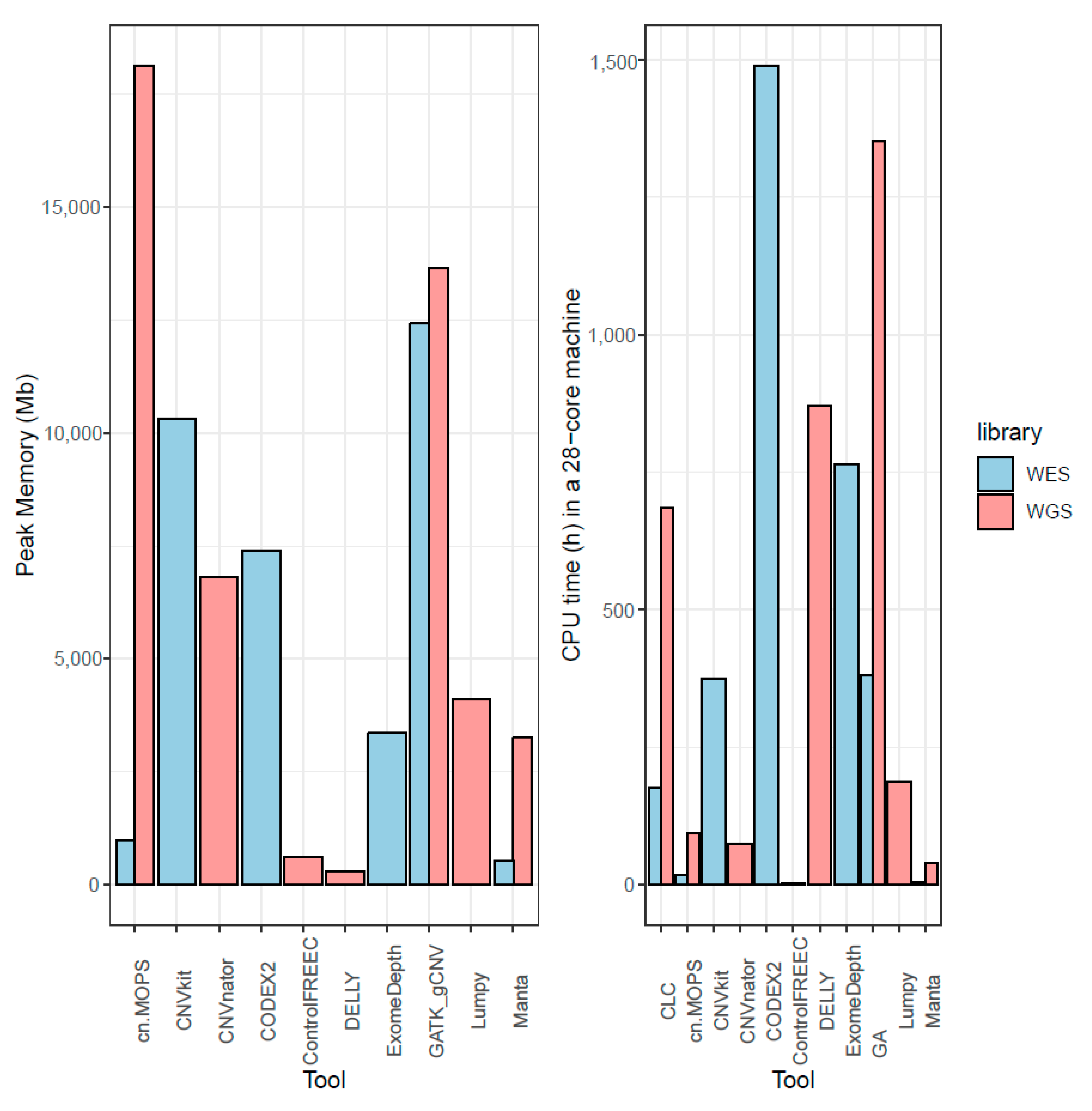
Cancers | Free Full-Text | A Comparison of Tools for Copy-Number Variation Detection in Germline Whole Exome and Whole Genome Sequencing Data

inCNV: An Integrated Analysis Tool for Copy Number Variation on Whole Exome Sequencing | Semantic Scholar

Comparative study of whole exome sequencing-based copy number variation detection tools | BMC Bioinformatics | Full Text
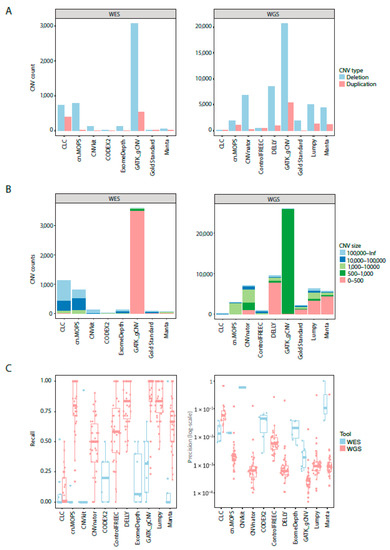
Cancers | Free Full-Text | A Comparison of Tools for Copy-Number Variation Detection in Germline Whole Exome and Whole Genome Sequencing Data
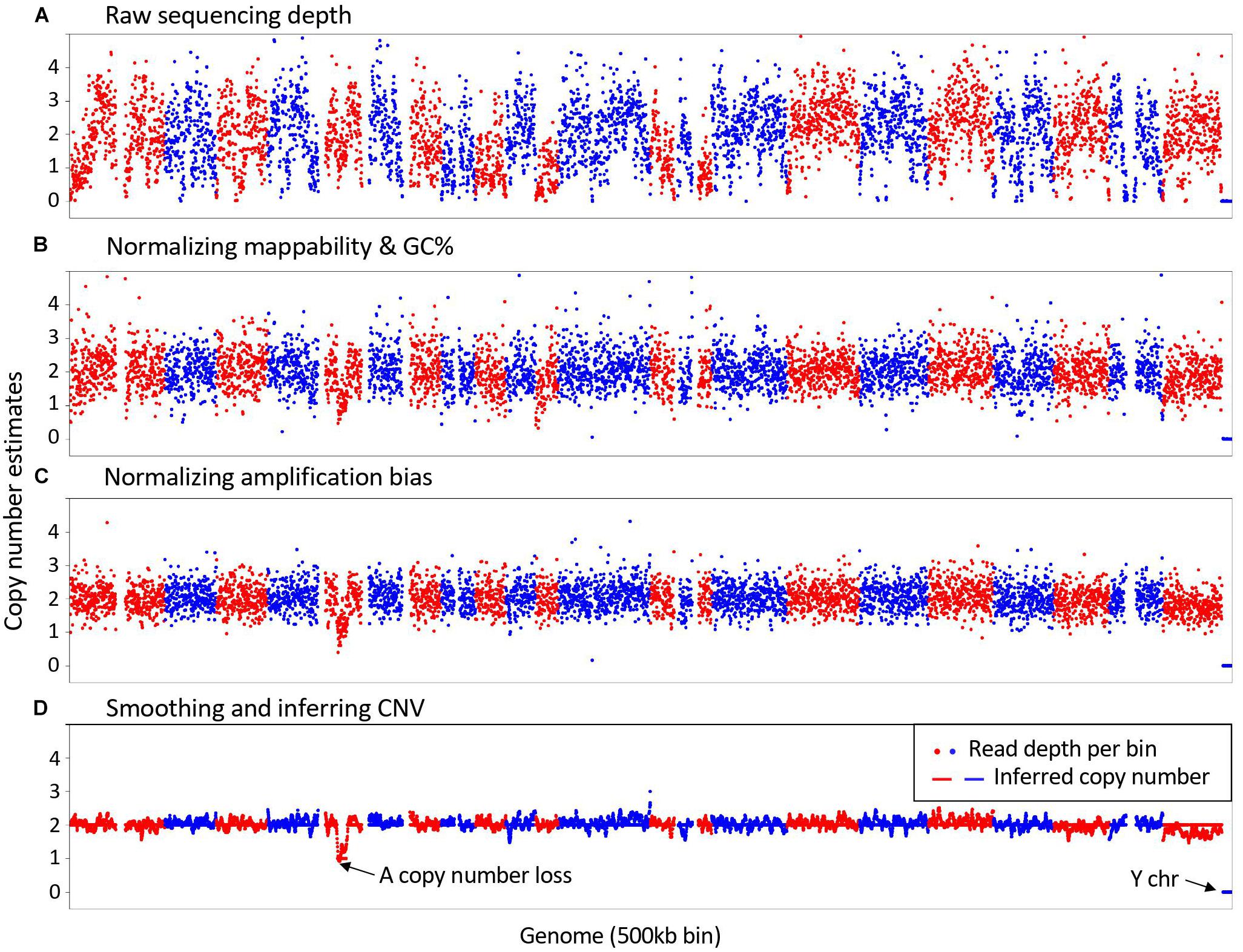
Frontiers | SCCNV: A Software Tool for Identifying Copy Number Variation From Single-Cell Whole-Genome Sequencing
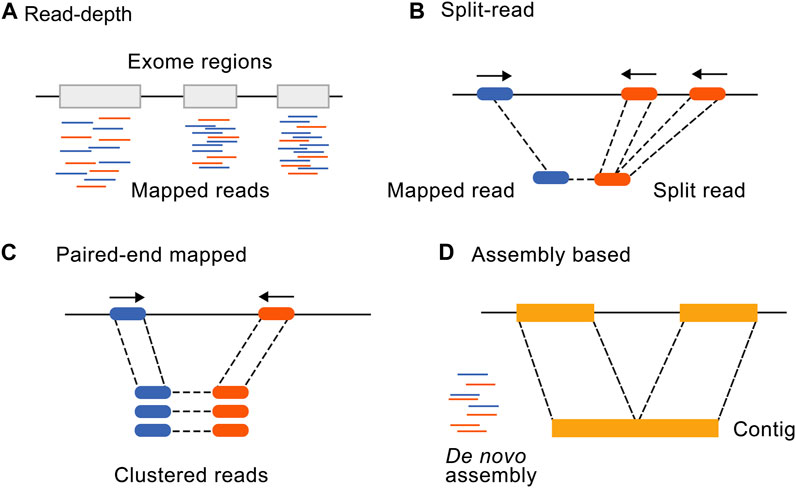
Frontiers | Incorporating CNV analysis improves the yield of exome sequencing for rare monogenic disorders—an important consideration for resource-constrained settings
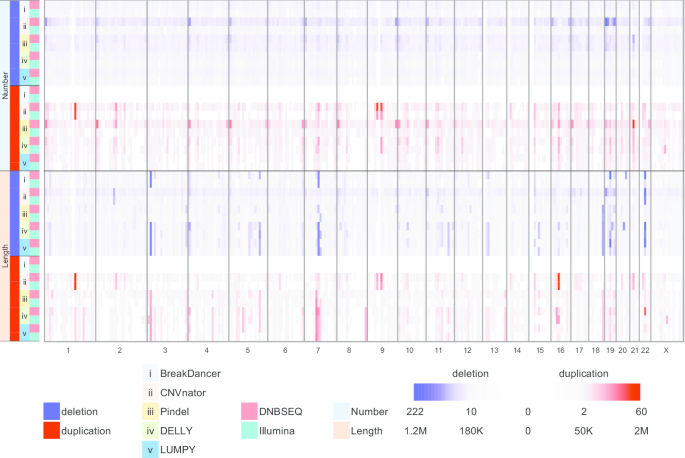
Performance of copy number variants detection based on whole-genome sequencing by DNBSEQ platforms | BMC Bioinformatics | Full Text